
TAX-Corpus: Taxonomy based Annotations for Colonoscopy
Evaluation
Shorabuddin Syed
1
a
, Adam Jackson Angel
2
, Hafsa Bareen Syeda
3
b
, Carole Franc Jennings
4
,
Joseph VanScoy
5
, Mahanazuddin Syed
1
c
, Melody Greer
1
, Sudeepa Bhattacharyya
6
,
Shaymaa Al-Shukri
1
, Meredith Zozus
7
d
, Fred Prior
1
e
and Benjamin Tharian
8
1
Department of Biomedical Informatics, University of Arkansas for Medical Sciences, U.S.A.
2
Department of Internal Medicine, Washington University, U.S.A.
3
Department of Neurology, University of Arkansas for Medical Sciences, U.S.A.
4
Department of Internal Medicine, Tulane University, U.S.A.
5
College of Medicine, University of Arkansas for Medical Sciences, U.S.A.
6
Department of Biological Sciences, Arkansas State University, U.S.A.
7
Department of Population Health Sciences, University of Texas Health Science Centre at San Antonio, U.S.A.
8
Division of Gastroenterology and Hepatology, University of Arkansas for Medical Sciences, U.S.A.
Keywords: Colonoscopy, Taxonomy, Annotation, Natural Language Processing, Machine Learning, Clinical Corpus.
Abstract: Colonoscopy plays a critical role in screening of colorectal carcinomas (CC). Unfortunately, the data related
to this procedure are stored in disparate documents, colonoscopy, pathology, and radiology reports
respectively. The lack of integrated standardized documentation is impeding accurate reporting of quality
metrics and clinical and translational research. Natural language processing (NLP) has been used as an
alternative to manual data abstraction. Performance of Machine Learning (ML) based NLP solutions is
heavily dependent on the accuracy of annotated corpora. Availability of large volume annotated corpora is
limited due to data privacy laws and the cost and effort required. In addition, the manual annotation process
is error-prone, making the lack of quality annotated corpora the largest bottleneck in deploying ML solutions.
The objective of this study is to identify clinical entities critical to colonoscopy quality, and build a high-
quality annotated corpus using domain specific taxonomies following standardized annotation guidelines. The
annotated corpus can be used to train ML models for a variety of downstream tasks.
1 INTRODUCTION
Colonoscopy plays a critical role in screening of
colorectal carcinomas (CC) (Kim et al., 2020).
Although it is a most frequently performed procedure,
the lack of standardized reporting is impeding clinical
and translational research. Vital details related to the
procedure are stored in disparate documents,
colonoscopy, pathology, and radiology reports
respectively. The established quality metrics such as
adenoma detection rates, bowel preparation, and
cecal intubation rate are documented in endoscopy
and pathology reports (Anderson & Butterly, 2015;
a
https://orcid.org/0000-0002-4761-5972
b
https://orcid.org/0000-0001-9752-4983
c
https://orcid.org/0000-0002-8978-1565
d
https://orcid.org/0000-0002-9332-1684
e
https://orcid.org/0000-0002-6314-5683
Rex et al., 2015). Procedure indicators, medical
history require review of clinical history and
radiology reports. A comprehensive study of quality
metrics often involves labour-intensive chart review,
thereby limiting the ability to report, monitor, and
ultimately improve procedure quality (Syed et al.,
2021).
Natural language processing (NLP) has been used
as an alternative to manual data abstraction. Previous
studies applied rule based or Machine Learning (ML)
based NLP solutions to extract limited clinical
concepts from unconsolidated procedure documents,
making the process of data extraction inadequate and
162
Syed, S., Angel, A., Syeda, H., Jennings, C., VanScoy, J., Syed, M., Greer, M., Bhattacharyya, S., Al-Shukri, S., Zozus, M., Prior, F. and Tharian, B.
TAX-Corpus: Taxonomy based Annotations for Colonoscopy Evaluation.
DOI: 10.5220/0010876100003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 5: HEALTHINF, pages 162-169
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

error-prone (Nayor, Borges, Goryachev, Gainer, &
Saltzman, 2018; Patterson, Forbush, Saini, Moser, &
DuVall, 2015). The ML algorithms are usually
trained and evaluated in the general English domain
and later applied to cross-domain settings (Lee et al.,
2019; Malte & Ratadiya, 2019; Schmidt, Marques,
Botti, & Marques, 2019). These off-the-shelf ML
models performs poorly on identifying clinical
concepts due to the lack of large annotated corpora
for training and the presence of domain specific
abbreviations and terminologies (Griffis, Shivade,
Fosler-Lussier, & Lai, 2016; Huang, Altosaar, &
Ranganath, 2019). There is a dearth of high-quality
annotated clinical corpora due to legal and
institutional concerns arising from the sensitivity of
clinical data (Spasic & Nenadic, 2020). Most
researchers are forced to build task specific annotated
corpora, which requires domain experts to review
hundreds of clinical narratives. The overall process of
manual annotation is both error-prone and expensive,
and considered as the largest bottleneck in deploying
ML solutions (Ratner et al., 2017). For colonoscopy
evaluation, this problem is exacerbated as procedure
metrics are distributed in multiple document types.
Several studies have been done to understand
factors effecting the annotation time and the quality
of clinical corpora (Fan et al., 2019; Roberts et al.,
2007; Wei, Franklin, Cohen, & Xu, 2018). Roberts et
al., 2007 (Roberts et al., 2007) and Wei et al., 2018
(Wei et al., 2018) identified number of entities to
annotate and long term dependencies between the
entities as the key factors hindering clinical text
annotations. Use of standard terminologies to
annotate clinical narratives reduces entity
identification ambiguities and improves syntactical
relation accuracies, allowing for high inter-annotator
agreement (Fan et al., 2019). Taxonomies facilitate
injecting domain knowledge into ML models and
improve clinical concept extraction accuracy (Jiang,
Sanger, & Liu, 2019; Wu et al., 2018). However,
colonoscopy documents are annotated to identify
specific procedure metrics and employing generic
terminologies will not be beneficial. To address this
problem, we built taxonomies specific to colonoscopy
documents and created a highly-accurate annotated
corpus based on the taxonomies and adoption of
standard annotation guidelines. This annotated corpus
can enhance ML model performance for downstream
tasks including clinical concept identification and
extraction.
2 METHODS
We built gold-standard annotated corpora using
colonoscopy documents of patients undergoing the
procedure at the University of Arkansas for Medical
Sciences (UAMS) from May 2014 to September
2020. As shown in Figure 1, the overall framework
for annotating colonoscopy related documents is
divided into two steps; 1) Pre-Annotation, and 2)
Interactive-Annotation. In the Pre-Annotation phase,
we identified clinical entities to annotate, build
taxonomies and annotation guidelines, recruited
annotators, and installed the annotation tool. In the
Interactive-Annotation phase, a random sample of
colonoscopy documents was selected and annotated
based on the provided guidelines. If the annotators
found any ambiguity or discovered new knowledge,
the taxonomies and annotation guidelines were
updated by the domain expert (BT).
2.1 Pre-annotation
From the three colonoscopy documents
(colonoscopy, pathology, and radiology reports), we
identified clinical entities that are essential to
measure, to improve procedure quality. To reduce
annotation time and improve quality of annotations,
we built taxonomies specific to each document type.
To identify clinical predictors from colonoscopy
procedure documents, we reviewed 15 quality metrics
published by the American College of
Gastroenterology and identified variables essential to
build the metrics (Anderson & Butterly, 2015; Rex et
al., 2015). We interviewed domain experts to
understand key factors leading to an aborted or
incomplete procedure (Brahmania et al., 2012). A
total of 74 entities embedded as free text in the
procedure report that can potentially impact
procedure outcome were selected for annotation. The
identified entities include scope times, quality of
bowel preparation, medications, type and location of
polyp etc. To build an annotation taxonomy specific
to colonoscopy reports, we classified the 74 entities
into 9 classes based on patient’s condition,
abnormalities found during the procedure, and
treatment plan. Figure 2 shows these classes and
associated entities.
To identify key pathological findings, we did an
extensive literature review on, 1) type of specimens
collected during the colonoscopy, and 2)
characteristic of polyps and their classifications. We
identified 61 principal entities that can be
documented in the reports and grouped these entities
into 4 classes: polyp type (neoplastic and non-
TAX-Corpus: Taxonomy based Annotations for Colonoscopy Evaluation
163
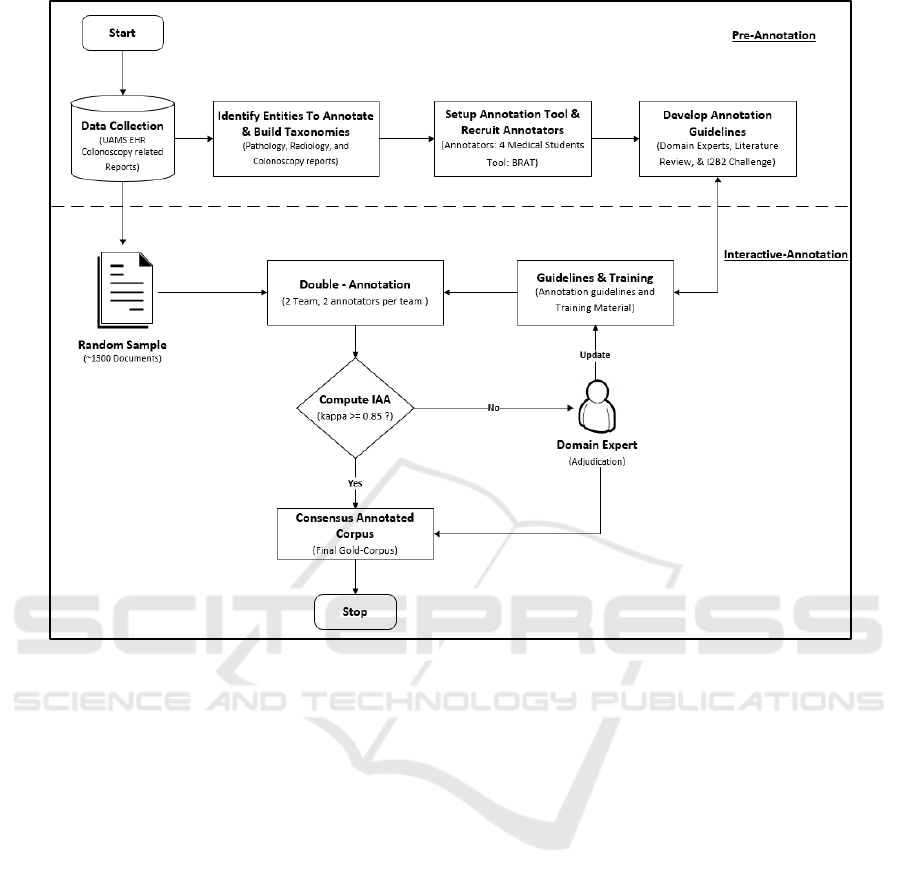
Figure 1: Annotation workflow to label colonoscopy related documents. The process is divided into pre-annotation and
interactive-annotation stage. In the pre-annotation stage clinical entities were identified, taxonomies created, annotators
recruited, and annotation guidelines and tools deployed. In the interactive-annotation stage, documents were double annotated
by two teams and differences was adjudicated by a domain expert.
neoplastic), pathological classification (benign and
malignant carcinomas), specimen type, and anatomic
location of the specimen. Figure 3 shows these classes
and associated entities.
To identify vital GI concepts from radiology
reports (i.e., Abdominal-pelvis CT scan, Abdominal
USG), a combination of manual review of the reports
and unsupervised topic modelling using Latent
Dirichlet Allocation (LDA) was done (Jelodar et al.,
2019). Based on the results from the aforementioned
methods, domain expert (BT) identified 47 entities.
These entities were classified into 2 primary classes,
abnormal findings and anatomical location. Figure 4
shows these classes and associated entities to annotate
from imaging reports.
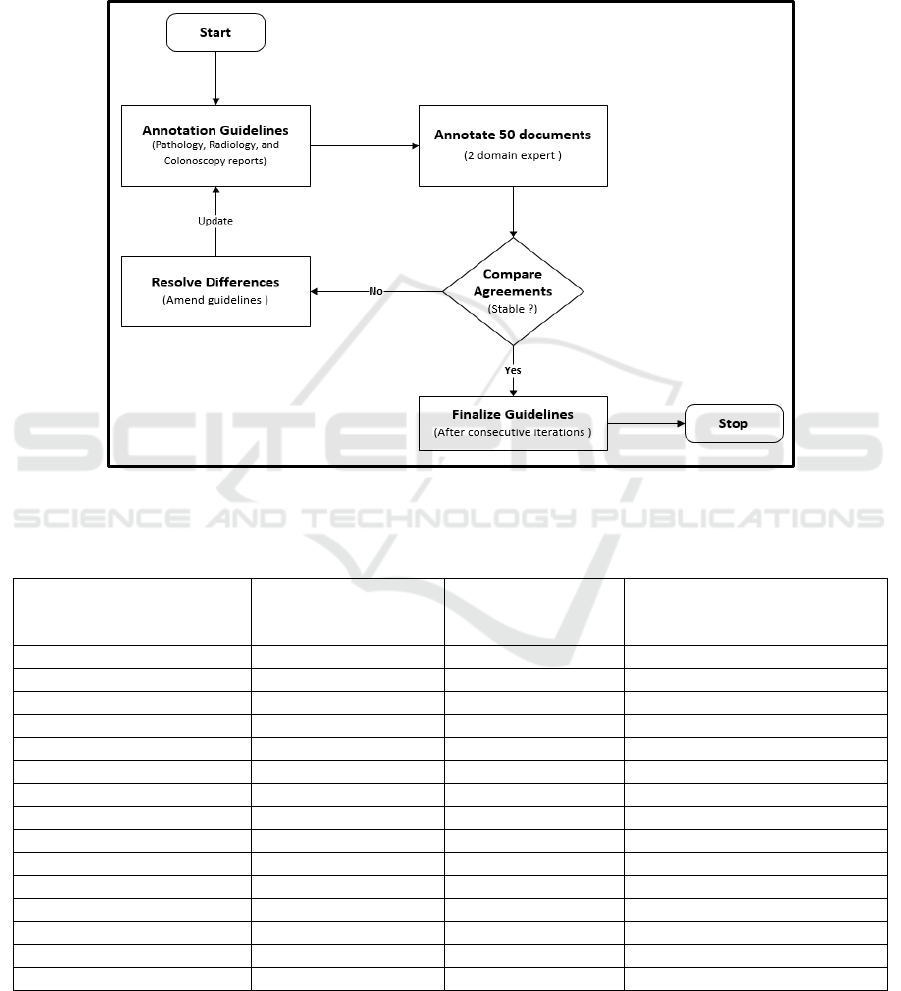
We drafted initial annotation guidelines based on
the guidelines published by Mehrotra et al., 2012 and
Informatics for Integrating Biology and Bedside
(i2b2) information extraction challenge workshops
(Henry, Buchan, Filannino, Stubbs, & Uzuner, 2020;
Sun, Rumshisky, & Uzuner, 2013). As shown in
Figure 5, the annotation guidelines were revised
through a rigorous and iterative process by two
qualified clinicians. Using the initial guidelines, a
random sample of 50 documents were annotated by
the clinicians in 5 iterations, the guidelines were
updated at the end of every iteration. We chose an
open-source annotation tool, BRAT (Stenetorp et al.,
2012) and recruited 4 medical students to annotate
procedure documents.
2.2 Interactive-annotation
A random sample of 1281 colonoscopy related
documents (Colonoscopy (CC) = 442, Pathology
(CP) = 426, Radiology (CR) = 413) were selected for
double annotation. Annotation guidelines was
provided to the annotators and training was provided
by the domain expert (BT). Two teams (2 students per
team) independently labelled the same set of
HEALTHINF 2022 - 15th International Conference on Health Informatics
164
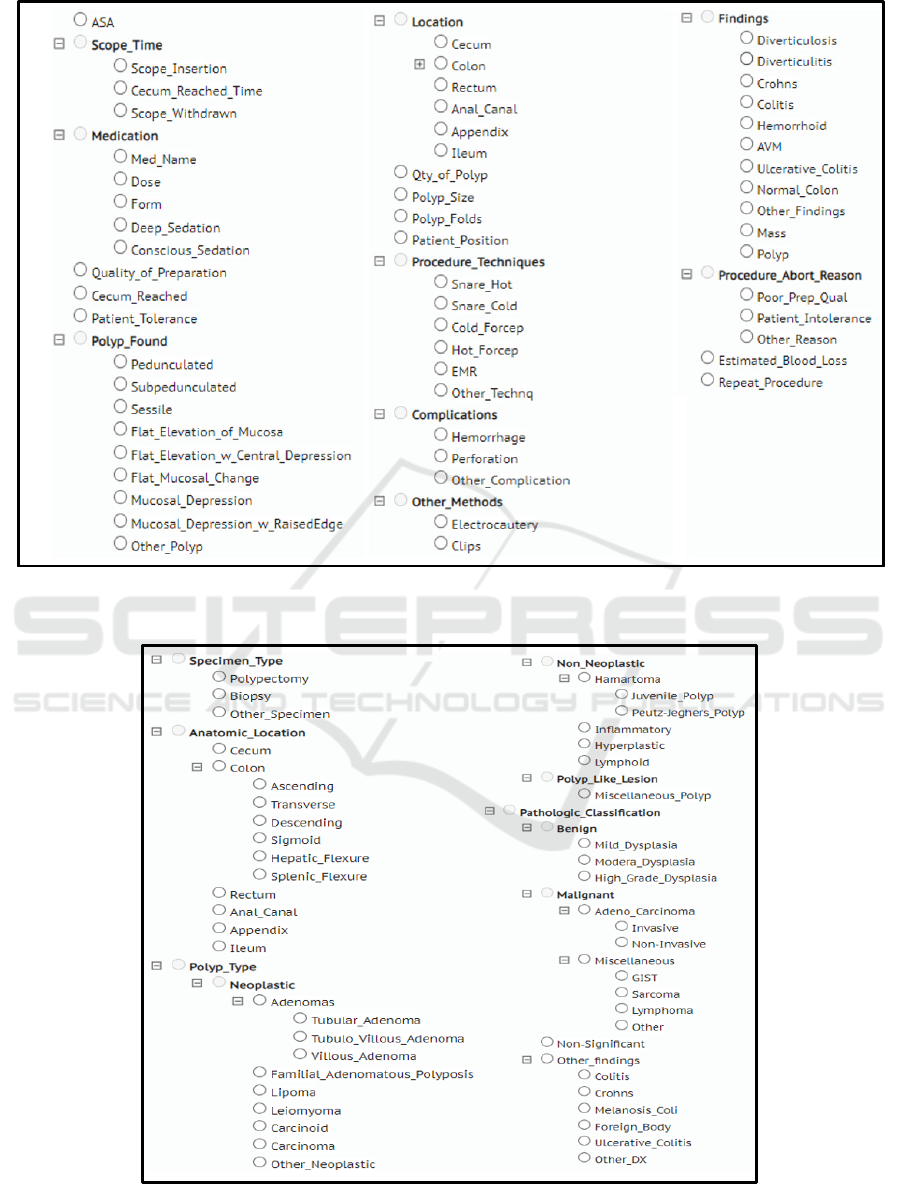
Figure 2: Colonoscopy taxonomy depicting clinical entities and their classifications. Colonoscopy reports were annotated for
entities mentioned in the taxonomy.
Figure 3: Pathology taxonomy depicting clinical entities and their classifications. Pathology reports were annotated for entities
mentioned in the taxonomy.
TAX-Corpus: Taxonomy based Annotations for Colonoscopy Evaluation
165

documents in three iterations, about 400 documents
per cycle. At the end of each iteration, we measured
agreement between the double annotated documents
using the inter annotator agreement (IAA) metric
shown in equation 1. Pairs of double annotations were
rejected if agreement did not pass the threshold (IAA
> 85%). In this case the domain expert adjudicated the
differences and re-trained the annotators. If new
knowledge was discovered during the process, the
annotation guideline was updated and occasionally
taxonomies were revised. The documents that passed
the set threshold were accepted into a consensus set.
The domain expert randomly reviewed documents
from this set to finalize the gold corpus.
IAA = matches / (matches + non-matches) (1)
Figure 4: Radiology imaging taxonomy depicting clinical
entities and their classifications. Radiology reports were
annotated for entities mentioned in the taxonomy.
3 RESULTS
Each annotator labelled 640 documents (1281 per
team) and took about 100-120 hours to complete the
annotation task. Of the three procedure documents,
colonoscopy reports had more entities to annotate (27
annotations for an average report). Table 1 shows the
total number of colonoscopy entities identified by
both teams from the annotated corpus. For pathology
and imaging documents, only final diagnosis and
impression sections of the reports were annotated
respectively. Table 2 shows the number of entities
identified by each team from the pathology reports.
For radiology reports, domain expert spent more time
adjudicating difference of agreements between the
annotators when compared to other report types. This
was due to complexity of the report and absence of
definitive conclusion. Table 3 shows the total number
of entities identified by each team from the radiology
reports. The final gold corpus consisted of 10,672,
4,136, and 3,071 entities from the colonoscopy,
pathology, and imaging reports respectively.
For every iteration, IAA between the annotators
improved. We believe that interactive discussion and
continuous training during the annotation process
helped achieve better IAA metrics. Overall IAA for
colonoscopy, pathology, and imaging reports was
0.910, 0.922, and 0.873 respectively.
4 DISCUSSION
Clinical narratives are often open to interpretation as
annotators use their own domain knowledge and
intuition to label the free text, thereby effecting the
annotation time and the quality of the resulting
corpus. Roberts et al., 2007 (Roberts et al., 2007) and
Wei et al., 2018 (Wei et al., 2018) identified various
factors hindering clinical text annotations: 1) intrinsic
characteristic of the documents, 2) annotator
expertise and annotation guidelines, 3) number of
entities to annotate, and 4) syntactical relations.
While building the 3 annotated corpora, we addressed
these concerns to a great extent. The annotation
guidelines including entities to annotate was drafted
by a panel of domain experts (GI physicians) based
on extensive literature review. The guidelines were
continuously updated based on new knowledge
discovered during the three annotation phases. The 4
annotators were highly qualified for the task, 3 of
them were final year medical students and 1 first year
internal medicine resident. To ensure consistency of
the annotated corpus, a domain expert trained the
annotators and annotation was strictly based on the
guidelines. As radiology reports are difficult to
interpret, domain experts allocated extra time to
evaluate and resolve annotation differences. To
minimize entity identification ambiguities and
improve syntactical relation accuracies, we injected
taxonomies specific to each document type. Double
annotation strategy and grouping related entities
under a single class, such as polyp types and removal
techniques, improved completeness and overall IAA
between the annotators.
HEALTHINF 2022 - 15th International Conference on Health Informatics
166
The annotation tool, BRAT was selected based on
extensive review of 15 tools done by Nevers et al.,
2019 (Neves & Ševa, 2021). BRAT was rated as the
most comprehensive and easy to use annotation tool,
and it supports normalization of pre-defined
terminologies. These features facilitated integration
of report-specific taxonomies for annotations.
Our study has several limitations. Abdominal CT
reports may contain findings from various vital
organs, for the scope of this project, we only
annotated conditions that can be diagnosed and
treated by GI endoscopy. Though the colonoscopy
reports were generated using multiple EHR software,
they were collected from a single institution (UAMS).
Figure 5: Workflow depicting development and refinement of annotation guidelines through a rigorous and iterative process.
Table 1: Clinical entities identified by two team of annotators from colonoscopy reports, and Inter Annotator Agreement
(IAA) between the teams.
Colonoscopy Entity Total Entities
Identified by Team 1
Total Entities
Identified by Team
2
Inter Annotator
Agreement (IAA) after 3
iterations
Scope_Time 1,310 1,290 0.981
Medication 2,112 2,214 0.913
Polyp_Found 505 509 0.962
Location 3,051 3,121 0.914
Procedure_Techniques 581 575 0.933
Findings 792 819 0.871
Complications 61 54 0.824
Other_Methods 206 227 0.911
Procedure_Abort_Reason 26 29 0.896
ASA 412 421 0.896
Cecum_Reached 398 402 0.951
Quality_of_Preparation 370 410 0.887
Patient_Tolerance 415 401 0.921
Estimated_Blood_Loss 325 349 0.917
Repeat_Procedure 37 33 0.860
TAX-Corpus: Taxonomy based Annotations for Colonoscopy Evaluation
167
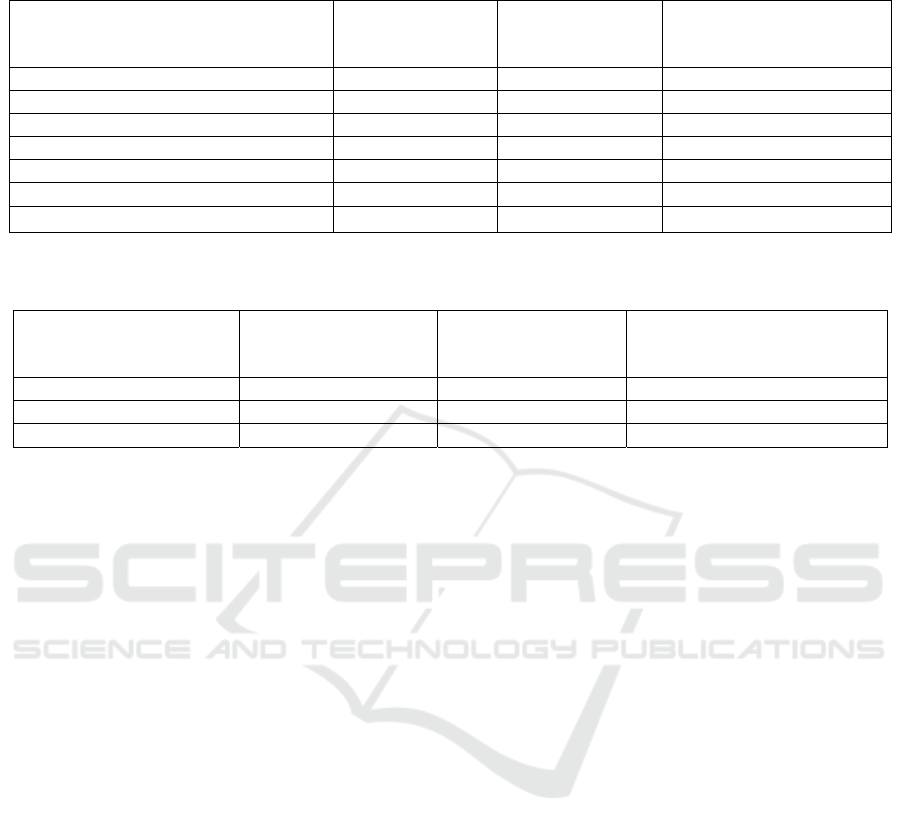
Table 2: Clinical entities identified by two team of annotators from pathology reports, and Inter Annotator Agreement (IAA)
between the teams.
Pathology Entity Total Entities
Identified by
Team 1
Total Entities
Identified by
Team 2
Inter Annotator
Agreement (IAA) after 3
iterations
Location 1,414 1,492 0.952
Specimen Type 1,330 1,371 0.948
Neoplastic Polyp 542 513 0.931
Non Neoplastic Polyp 396 317 0.897
Polyp Like Lesion 77 86 0.934
Pathological Classification Benign 285 306 0.893
Pathological Classification Malignant 93 81 0.868
Table 3: Clinical entities identified by two team of annotators from imaging reports, and Inter Annotator Agreement (IAA)
between the teams.
Imaging Entity Total Entities
Identified by Team 1
Total Entities
Identified by Team 2
Inter Annotator
Agreement (IAA) after 3
iterations
Findings 1,350 1,408 0.873
Location 1,431 1,364 0.882
Miscellaneous 192 213 0.865
5 CONCLUSION
Training data quality, both accuracy and consistency
of annotations, plays a crucial role in ML model
performance and evaluation. Using domain specific
taxonomies and adopting standard annotation
guidelines, we built high-quality colonoscopy corpus
needed to train ML models. The trained model can
then be used for downstream task of clinical concept
extraction crucial to colonoscopy quality evaluation.
Automated and accurate extraction of procedure
metrics will significantly reduce data accessibility
time and facilitates clinical and translational
endoscopy research.
ACKNOWLEDGMENT
Patient’s data used were obtained under IRB approval
(IRB# 262202) at the UAMS. This study was
supported in part by the Translational Research
Institute (TRI), grant UL1 TR003107 received from
the National Center for Advancing Translational
Sciences of the National Institutes of Health (NIH).
The content of this manuscript is solely the
responsibility of the authors and does not necessarily
represent the official views of the NIH.
REFERENCES
Anderson, J. C., & Butterly, L. F. (2015). Colonoscopy:
quality indicators. Clinical and translational
gastroenterology, 6(2), e77-e77.
doi:10.1038/ctg.2015.5
Brahmania, M., Park, J., Svarta, S., Tong, J., Kwok, R., &
Enns, R. (2012). Incomplete colonoscopy: maximizing
completion rates of gastroenterologists. Canadian
journal of gastroenterology = Journal canadien de
gastroenterologie, 26(9), 589-592.
doi:10.1155/2012/353457
Fan, Y., Wen, A., Shen, F., Sohn, S., Liu, H., & Wang, L.
(2019). Evaluating the Impact of Dictionary Updates on
Automatic Annotations Based on Clinical NLP
Systems. AMIA Jt Summits Transl Sci Proc, 2019, 714-
721.
Griffis, D., Shivade, C., Fosler-Lussier, E., & Lai, A. M.
(2016). A Quantitative and Qualitative Evaluation of
Sentence Boundary Detection for the Clinical Domain.
AMIA Jt Summits Transl Sci Proc, 2016, 88-97.
Henry, S., Buchan, K., Filannino, M., Stubbs, A., &
Uzuner, O. (2020). 2018 n2c2 shared task on adverse
drug events and medication extraction in electronic
health records. J Am Med Inform Assoc, 27(1), 3-12.
doi:10.1093/jamia/ocz166
Huang, K., Altosaar, J., & Ranganath, R. (2019).
ClinicalBERT: Modeling Clinical Notes and Predicting
Hospital Readmission.
Jelodar, H., Wang, Y., Yuan, C., Feng, X., Jiang, X., Li, Y.,
& Zhao, L. (2019). Latent Dirichlet allocation (LDA)
and topic modeling: models, applications, a survey.
HEALTHINF 2022 - 15th International Conference on Health Informatics
168

Multimedia Tools and Applications, 78(11), 15169-
15211. doi:10.1007/s11042-018-6894-4
Jiang, M., Sanger, T., & Liu, X. (2019). Combining
Contextualized Embeddings and Prior Knowledge for
Clinical Named Entity Recognition: Evaluation Study.
JMIR Med Inform, 7(4), e14850. doi:10.2196/14850
Kim, K., Polite, B., Hedeker, D., Liebovitz, D., Randal, F.,
Jayaprakash, M., . . . Lam, H. (2020). Implementing a
multilevel intervention to accelerate colorectal cancer
screening and follow-up in federally qualified health
centers using a stepped wedge design: a study protocol.
Implementation Science, 15(1), 96.
doi:10.1186/s13012-020-01045-4
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., &
Kang, J. (2019). BioBERT: a pre-trained biomedical
language representation model for biomedical text
mining. Bioinformatics (Oxford, England).
doi:10.1093/bioinformatics/btz682
Malte, A., & Ratadiya, P. (2019). Evolution of transfer
learning in natural language processing. CoRR,
abs/1910.07370.
Nayor, J., Borges, L. F., Goryachev, S., Gainer, V. S., &
Saltzman, J. R. (2018). Natural Language Processing
Accurately Calculates Adenoma and Sessile Serrated
Polyp Detection Rates. Dig Dis Sci, 63(7), 1794-1800.
doi:10.1007/s10620-018-5078-4
Neves, M., & Ševa, J. (2021). An extensive review of tools
for manual annotation of documents. Brief Bioinform,
22(1), 146-163. doi:10.1093/bib/bbz130
Patterson, O. V., Forbush, T. B., Saini, S. D., Moser, S. E.,
& DuVall, S. L. (2015). Classifying the Indication for
Colonoscopy Procedures: A Comparison of NLP
Approaches in a Diverse National Healthcare System.
Stud Health Technol Inform, 216, 614-618.
Ratner, A., Bach, S. H., Ehrenberg, H., Fries, J., Wu, S., &
Re, C. (2017). Snorkel: Rapid Training Data Creation
with Weak Supervision. Proceedings VLDB
Endowment, 11(3), 269-282.
doi:10.14778/3157794.3157797
Rex, D. K., Schoenfeld, P. S., Cohen, J., Pike, I. M., Adler,
D. G., Fennerty, M. B., . . . Weinberg, D. S. (2015).
Quality indicators for colonoscopy. Gastrointest
Endosc, 81(1), 31-53. doi:10.1016/j.gie.2014.07.058
Roberts, A., Gaizauskas, R., Hepple, M., Davis, N.,
Demetriou, G., Guo, Y., . . . Wheeldin, B. (2007). The
CLEF corpus: semantic annotation of clinical text.
AMIA ... Annual Symposium proceedings. AMIA
Symposium, 2007, 625-629.
Schmidt, J., Marques, M. R. G., Botti, S., & Marques, M.
A. L. (2019). Recent advances and applications of
machine learning in solid-state materials science. npj
Computational Materials, 5(1), 83.
doi:10.1038/s41524-019-0221-0
Spasic, I., & Nenadic, G. (2020). Clinical Text Data in
Machine Learning: Systematic Review. JMIR Med
Inform, 8(3), e17984-e17984. doi:10.2196/17984
Stenetorp, P., Pyysalo, S., Topić, G., Ohta, T., Ananiadou,
S., & Tsujii, J. i. (2012, apr). brat: a Web-based Tool
for NLP-Assisted Text Annotation, Avignon, France.
Sun, W., Rumshisky, A., & Uzuner, O. (2013). Evaluating
temporal relations in clinical text: 2012 i2b2 Challenge.
J Am Med Inform Assoc, 20(5), 806-813.
doi:10.1136/amiajnl-2013-001628
Syed, S., Tharian, B., Syeda, H. B., Zozus, M., Greer, M.
L., Bhattacharyya, S., . . . Prior, F. (2021). Consolidated
EHR Workflow for Endoscopy Quality Reporting. Stud
Health Technol Inform, 281, 427-431.
doi:10.3233/shti210194
Wei, Q., Franklin, A., Cohen, T., & Xu, H. (2018). Clinical
text annotation - what factors are associated with the
cost of time? AMIA Annu Symp Proc, 2018, 1552-1560.
Wu, Y., Yang, X., Bian, J., Guo, Y., Xu, H., & Hogan, W.
(2018). Combine Factual Medical Knowledge and
Distributed Word Representation to Improve Clinical
Named Entity Recognition. AMIA Annu Symp Proc,
2018, 1110-1117.
TAX-Corpus: Taxonomy based Annotations for Colonoscopy Evaluation
169
