
microCT for Systematic Mouse Phenotyping
Frantisek Spoutil, Michaela Prochazkova, Tereza Michalcikova, Ivana Uramova, Sarah Clewell,
Vendula Novosadova and Jan Prochazka
Czech Centre for Phenogenomics, BIOCEV - IMG, Prumyslova 595, Vestec, Czech Republic
Keywords: 3D Imaging, Animal Model, Body Composition, Bone Density, Embryology, Machine Learning,
Morphology, Mus.
Abstract: Phenotyping of mouse mutants is one of the crucial methods for uncovering genetic network at the level of a
whole organism which could help us to understand origin of rare diseases, developmental malformations, but
also the process of mammalian evolution. For studying morphological aspects of either embryos or adults, the
X-ray computed microtomography (microCT) has become a gold standard within the last years. The three-
dimensional (3D) context, availability of data to additional analysis (e.g. volumetric, bone density, or body
composition), and in-vivo approaches in the case of adults are the main advantages when compared to classic
histology and bone morphology. On the other hand, the amount of data is enormous making the data storage
and analysis the bottle-neck of the microCT method. To overcome this obstacle, we cooperate with
bioinformatics experts to set up automation of the process at maximal possible level. Nevertheless, knowledge
and experience of a specialist remain indispensable.
1 INTRODUCTION
The aim to understand and fully annotate mammalian
genome led to establishment of the International
Mouse Phenotype Consortium (IMPC) for the
systematic generation and analysis of all coding
sequence mutations in mice. On the basis of IMPC,
we have built an advanced phenotyping pipeline
using in-vivo microCT scanning technology for adult
mutant mouse cohorts. The primary challenge of
implementing 3D phenotyping of embryos using
microCT technology was to create standardization
across all stages of embryonic development.
However, the advanced, high resolution microCT
scanning allows for detailed morphological analysis
of embryos from the earliest developmental stages up
to perinatal period. The standardization of the whole
process, as established by IMPC, is critical for
relevant comparison and reproducibility of data
between research centers.
The advantages of 3D data generation compared
to more conservative approaches, such as plain X-ray
imaging, for adult skeletons or histological analysis
of embryos are obvious. While the products of
microCT imaging are incredibly detailed, the system
requires significant and time-consuming efforts in
order to process and further analyse the large 3D
datasets. This article will provide an overview of the
standardised morphological phenotyping pipeline, as
well as outline the data analysis in further detail.
Here we present the contemporary, state-of-the-
art high-throughput embryo and bone morphology
pipelines of mouse phenotyping used in our
department with the help of microCT technology, as
well as the obstacles we try to solve to reduce the
disadvantages of the method. We hope this approach
can make the microCT method more accessible to
broader spectrum of researchers, better their results
significantly, and reduce amount of animals used in
the experiments in the future.
2 METHODS
2.1 Embryology
The standardised embryo phenotyping pipeline
contains multiple critical steps for reproducibility and
reliability of data: proper and unified embryo
breeding, embryo harvest and fixation, contrasting of
soft tissues, microCT scanning with metadata
recording, data reconstruction and processing, data
upload to public open database mousephenotype.org.
144
Spoutil, F., Prochazkova, M., Michalcikova, T., Uramova, I., Clewell, S., Novosadova, V. and Prochazka, J.
microCT for Systematic Mouse Phenotyping.
DOI: 10.5220/0007570701440149
In Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2019), pages 144-149
ISBN: 978-989-758-353-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

2.1.1 Embryo Phenotyping Workflow
To determine the effects of embryonic lethal gene
mutations on development, embryos are harvested
from pregnant females a strictly systematic way,
leading to identification and characterisation of lethal
phenotypes. The initial stage for investigation is
E12.5. In case there are no living knockout (KO)
embryos at this stage the E9.5 embryos are analysed.
If KO embryos are absent at E9.5, earlier
developmental stages are then analysed individually.
In case there are living KO embryos at E12.5, the
subsequent developmental stages (E14.5 and E18.5)
are analysed.
After mating, females are visually examined
every morning for a presence of the vaginal plug,
which indicates embryonic day 0.5 (E0.5) of
development.
Gravidity is confirmed by a weight gain or at
earlier stages by ultrasonography. During embryo
harvest on the desired day of embryonic
development, yolk sacks are collected for genotyping
and embryos are fixed with 4% paraformaldehyde
(PFA).
2.1.2 Embryo Contrasting
High-resolution microCT provides an opportunity to
visualize embryos at various stages of development in
3D. Due to weak tissue mineralisation, a contrast
agent must be applied to all specimens.
Smaller samples, e.g. E9.5 embryos, are fixed for
24 hours in 4% PFA and stained with 1% PTA
(phosphotungstic acid) for up to 2 weeks. Larger
samples, e.g. E18.5 embryos, are fixed for 1 week in
4% PFA and stained with Lugol’s Iodine solution for
2 weeks or longer. Lugol’s stock solution (10g KI and
5g I
2
in 100ml H
2
O;) is diluted to 25% working
solution in H
2
O to achieve neutral osmotic pressure
to avoid tissue distortion.
2.1.3 Embryo Scanning and Reconstruction
Stained specimens are removed from the contrast
agent, rinsed with PBS and embedded in 2.5% low-
gelling temperature agarose in tubes. Tubes of
various sizes are used, depending on embryo size,
(single 0.2ml PCR microtubes, 2ml microtubes with
caps or 15ml falcon tubes cut to desired length). All
specimens have to be wrapped in Parafilm to prevent
evaporation.
Depending on the embryo size, SkyScan 1272
high-resolution microCT (Bruker, Belgium) is set up
for voxel size 0.2 - 7µm, and 0.5 or 1 mm Al filter.
360° scan with 0.200° rotation step and 3 frames
averaging setup is used for scanning. Scanning takes
from 5 to 20 hours per one sample, depending on size.
Automated scanning of multiple samples is acquired
by using sample carousel.
InstaRecon CBR Premium software (InstaRecon,
USA) is used for reconstruction. The setup of
reconstruction parameters such as smoothing, ring
artefacts correction, beam hardening and intensities
depends on the embryonic stage.
2.1.4 Embryo Phenotyping
Within the embryonic lethal screen, three knockout
embryos and one littermate, wild-type embryo are
scanned and their phenotypes are evaluated. Gross
morphology (growth retardation, development of
limbs, formation of orofacial area, etc.) is assessed in
whole-mount images and defects of inner organs
(positioning and size of organs, tooth development,
presence of cleft palate, etc.) are examined in the
virtual sections.
2.2 Adult Morphology
For standard morphological phenotyping, a cohort of
28 mice (14 wild types and 14 mutants, composed
from 7 males and 7 females each) at the age of 13
weeks is scanned in-vivo.
2.2.1 Adult Morphology Workflow
Each mouse is anesthetized by Zoletile injection,
arranged in natural position and scanned in SkyScan
1176 in-vivo microCT (Bruker, Belgium). After
scanning, the mouse is weighed and some basic body
measurements are obtained with digital dial calliper.
Two reconstructions are produced from the
primary data: i) for skeletal morphology and bone
mineral density (BMD), and ii) for body composition
analysis.
While the bone morphology is evaluated directly
from its reconstruction files, for body composition
analysis the core body is selected first before entering
the analysis. Volume of interest (VOI) from body
composition analysis is used also for BMD analysis
excluding skull, tail and distal limbs.
The whole process takes about 34 working hours
and 0.314 TB per cohort. See Figure 1 for a detail.
2.2.2 Mouse Scanning and Reconstruction
SkyScan 1176 in-vivo microCT (Bruker, Belgium) is
set up for voxel size 35 µm, voltage of 50 kV, current
of 160 µA, and 0.5 mm aluminium filter with 180°
rotation. When using these parameters, scanning one
microCT for Systematic Mouse Phenotyping
145

Figure 1: Scheme of adult mouse morphology and body composition workflow. Black: data acquisition; Red: data
reconstruction; Orange: data preparation; Green: analysis; Blue: data storage; Grey: Excel output. smooth. = smoothing; ring
art. corr. = ring artifact correction; hard. = beam hardening; def. px. mask = defect pixel masking; CS = border intensities for
reconstruction.
mouse takes about 16 minutes and the mouse takes
about 55.4 mGy/min.
For reconstruction InstaRecon CBR Premium
software (InstaRecon, USA) is used. In the case of
bone morphology, the reconstruction parameters, as
recommended by Bruker microCT (Belgium) are set
up as follows: smoothing = 3, ring artefact correction
= 4, beam hardening = 36%, intensities = 0.0047 –
0.1230. For body composition analysis the values are
changed to: smoothing = 7, ring artefacts correction =
= 5, beam hardening = 10%, defect pixel masking =
5%, intensities = 0.0040 – 0.0200.
2.2.3 Bone Morphology Phenotyping
CTvox software (Bruker microCT, Belgium) is the
basic tool for bone morphology evaluation used. We
record 53 qualitative and numerical variables
describing axial, brachial, and cranial skeleton to
localize effect of the mutation. 8 standardized views
on the whole skeleton or its selected parts (cranium,
limbs) are taken for each sample, as well as details of
malformations if they occur.
2.2.4 Body Composition Analysis
CT analyzer (CTan: Bruker microCT, Belgium) and
Batch Manager (BatMan: Bruker microCT, Belgium)
software are used for VOI selection as well as
analysis itself.
The analysis is based on different density of bones
(and teeth), lean, and fat (with lungs) which is then
distinguishable by X-rays. Primary data includes
absolute and relative volume of all three parts, which
are recalculated to absolute and relative mass
assumptions of lean and fat.
Machine learning-based procedure is developed
and tested at our centre to automatize the VOI
selection.
2.2.5 Bone Mineral Density
The same software is used also for evaluation of bone
mineral density (BMD) and tissue mineral density
(TMD) within the VOI of body composition analysis.
The values are calculated based on results from
calibrated hydroxyapatite phantoms scanned and
reconstructed under the same conditions.
Two secondary variables are computed from the
basic ones: bone mineral content (BMC), and BMC
per body mass.
2.3 Bioinformation Solution
As one of the primary goals in phenotyping is to
implement a high-throughput approach, maximum
automation of the whole procedure with the help of
machine-learning is optimal for our methodological
development, now and in the future. Although we
want to end up with total automation, we start step by
step by solving the most time-demanding problems,
which are i) VOI selection and ii) extraction of
analysis results, for body composition analysis.
Manual selection in CTan was originally used for
VOI definition. The biggest challenge for automation
is how to train the software to delineate what is the
BIOIMAGING 2019 - 6th International Conference on Bioimaging
146
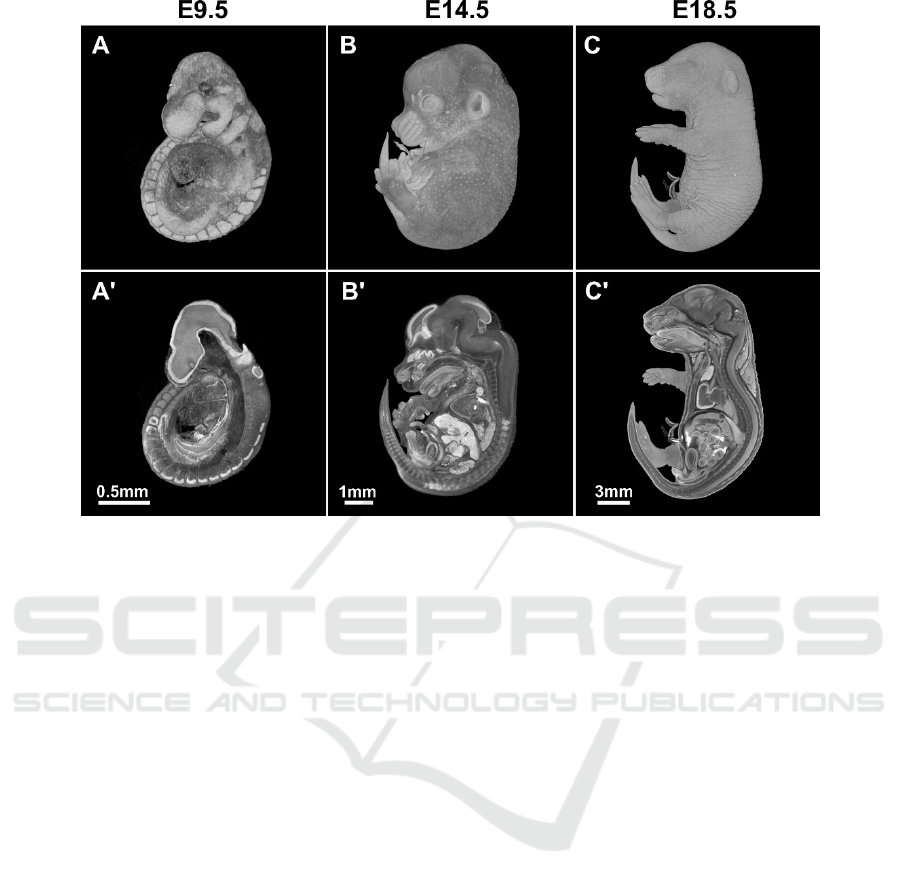
Figure 2: MicroCT scans of wild type murine embryos at different stages of development. A-C - whole-mount views, A'-C' -
virtual sagittal sections.
relevant specimen (i.e. mouse), as the size, shape, and
even orientation of some parts (e.g. limbs) have
significant variability between samples and slices. To
overcome this huge variability, machine-learning
using U-net deep neural network in PyTorch (Paszke
et al., 2017) with principal component analysis based
reorientation in R (R Core Team, 2015) and RStudio
(RStudio Team, 2016) with EBImage (Pau et al.,
2010), pixmaps (Bivand et al., 2015), and magick
(Ooms, 2018) libraries are applied based on previous
manual selection. Although the algorithm is still
under development and results need to be check by a
specialist, it has already increased the efficiency of
VOI selection 15:1 hour per whole mouse cohort (i.e.
28 mice). The data for VOI selection is rotated first
with principal component analysis (PCA), the starting
virtual section image for VOI is selected based on the
shape similarity in the cervical area, and then the
software crops all images section by section, i.e. in
2D to keep core body only. The accuracy of selecting
the 1
st
virtual slice is very high and differs a few slides
from selection of specialist, especially in animals
with less standard position (e.g. when shoulders are
moved more cranially). We were testing also VOI
selection based on 3D model, but the results were
comparable to 2D model approach, but computation
time was longer. Moreover, 2D approach enables
better correction of results by specialist. While
training the algorithm for automatic selection of the
areas of interest, we used 100 mice (more than 60
thousands virtual sections), which were manually
corrected. The final algorithm has been now used for
more than 1000 mice, and will be retrained to achieve
increasingly higher accuracy.
CTan saves results of every analysis to .CSV table
format with all procedures data acquired. The ideal
final stage of automation would connect the ROI
selection and data extraction with CTan’s macro for
body composition analysis along with its
sophisticated methods for smoothing, noise reduction
and separation.
However, the real challenge is automation of bone
morphology. We want the software to be able to
distinguish and identify individual bones or parts of
the skeleton (e.g. spine, skull, paws), to compare them
with standard shape variability of baseline, and
highlight any differences worthy of attention of a
morphologist. Although some similar approaches
already exist (e.g. Baiker et al., 2010, Wise et al.,
2013), their results are not fully applicable for our
demands and their optimization or finding of original
solution will be needed.
microCT for Systematic Mouse Phenotyping
147

3 RESULTS & DISCUSSION
3.1 Embryo Morphology
Within the embryology lethal screen, we have at the
moment phenotyped 12 lineages. One of the lineages
was lethal before E9.5, four were perinatal lethal and
two were identified as “subviable” lineages. The rest
of strains were lethal between E9.5 and birth. See
Figure 2 for examples of scans and virtual sections of
wild type embryos at different developmental stages.
For embryos older than E9.5, we used the Lugol’s
solution as contrast agent. Noteworthy, dilution of
working solution in water and not PBS turned out to
be methodologically crucial as this approach does not
cause tissue shrinkage.
We have observed various pathologies, such as
growth retardation, short face, and heart and intestinal
dysmorphology in the embryos. We could evaluate
the latter mentioned phenotypes thanks to the high
resolution microCT scanning. It would be very
difficult or even impossible to get this data from
classical histology, especially in cases where we
assessed the length and shape of the inner organs (e.g.
in case of embryonic intestine).
Our next goal in embryo screen is the adoption of
an atlas-based approach of organ recognition shared
among the IMPC centers (eg. Brown et al., 2018),
which will point out even slighter differences in organ
shape and position to a researcher, and quantify the
volume.
3.2 Adult Morphology
In-vivo microCT use in adult mouse morphology
brings numerous advantages compared to standard,
2D X-ray imaging: the level of detail is much higher,
we are able to select appropriate angle and section of
view to show a structure of interest without
compromise from X-ray shielding by surrounding
tissues. In that way we are able to observe structures
like rib rudiments on cervical vertebra, even slight
opening of skull sutures or dorsal arches of vertebrae,
occurrence of baculum in females, or ossification in
tendons of mouse. See Figure 3. This results in higher
probability an effect of mutation will be uncovered
(especially in heterozygotes) and that we will be able
to separate it from general genetic background of the
mouse strain. We have scanned and analysed 27,825
WT mice of both sexes so far, which serves as
baseline of comparison for the relevance of abnormal
morphology findings in KO mutant mice. There were
784 WT mice (2.74 %) with some abnormal
morphology findings. This number is almost two
times lower than in KO mutant mice (all mutations
together), where some abnormality was recorded in
1680 animals from 33,263 (4.81 %).
The greatest advantage of microCT though, is the
possibility to reuse the data of spatially different X-
ray absorption repeatedly. This quality is used also for
body composition analysis, where double-energy X-
ray analysis (DXA) is standard for IMPC. In the case
of DXA the amount of fat and lean is computed from
differences in absorption of X-rays of two energies.
MicroCT brings another quality: spatial distribution
of fat and lean, which is important especially for fat
tissue, as differences there are clinical differences,
whether the fat is stored more subcutaneously or
viscerally. In our department, significant differences
in baseline WT body composition were found in 17
of 46 KO mutant mouse gene cohorts with this
analysis so far.
Figure 3: Examples of skeletal dysmorphology. A) Rib
osteoneogenesis due to injuries in mouse mutant (compare
to B and D). B) Malformation of scapula and ulna in mouse
mutant (compare to A). C) Malformation of the 1
st
and 2
nd
thoracic vertebra in wild type mouse: 1
st
one without fusion
of dorsal arch, 2
nd
one without prolongation of spinal
process (compare to D). D) Cervical rib of 7
th
cervical
vertebra causing malformation of the 1
st
thoracic rib in
mouse mutant. E) Extra ossification in pelvic region of
mouse mutant (compare to F). F) 6
th
lumbar vertebra with
morphology of the 1
st
sacral one in mouse mutant (compare
to E). S1: 1
st
sacral vertebra; T1: 1
st
thoracic vertebra;
arrowheads: pointing to mentioned malformation. Figures
not in the same scale.
Magnetic resonance imaging (MRI) can be used
for body composition and even for general skeletal
morphology too. Its results are even much better for
soft tissue morphology. However, the crucial
disadvantage (besides much higher working costs
compared to microCT) is, that it isn’t able to quantify
bone density.
BIOIMAGING 2019 - 6th International Conference on Bioimaging
148

In the case of microCT, its computational and
time requirements are as summarized in Figure 1.
Increasing of computational power and machine
learning programming, which we are working on and
which was summarised elsewhere (Spoutil et al.,
2018), we will be able to push usability of microCT
for standard phenotyping procedure to broaden the
spectrum of usage and users. In the case of body
composition, the next goal is to teach the software to
differentiate hard particles of food from bone and
remove them from sections, plus smooth artificially-
increased intensities in their surroundings causing
star-like artefacts, which can distort real borders of fat
and lean, and thus their estimated volume. In the case
of bone morphology, we are planning to use a 3D
atlas-based approach similar to embryo screen (e.g.
Baiker et al., 2010) able to highlight significant
changes from mean morphology of individual bones,
as well as sections of skeleton.
We have clearly demonstrated that the data
quality of our approach is equal or higher than in the
standard 2D methods used in descriptive morphology
and anatomy of embryos and adults of mice due to
lower tissue deformation, full 3D spatial context, re-
usability of data etc. Replacing the work of specialists
with machine-learning and automation of the
procedure is the way to overcome the biggest
disadvantage of the method time demands. Its
application brought us first significant time savings.
Nevertheless, we still believe, the main role of the
computers in this process should be to help
researchers to focus more on data of their interest,
instead of fully automatic analysis. This is the way we
want to direct our future development of our
procedure.
ACKNOWLEDGEMENTS
This work was supported by RVO 68378050 by the
Academy of Sciences of the Czech Republic,
LM2015040 Czech Centre for Phenogenomics by
MEYS, CZ.02.1.01/0.0/0.0/16_013/0001789
Upgrade of the Czech Centre for Phenogenomics:
developing towards translation research by MEYS
and ERDF, CZ.1.05/2.1.00/19.0395 Higher quality
and capacity for transgenic models by MEYS and
ERDF, and CZ.1.05/1.1.00/02.0109 Biotechnology
and Biomedicine Centre of the Academy of Sciences
and Charles University in Vestec (BIOCEV) by
MEYS and ERDF. We also want thank to Radislav
Sedlacek, director of CCP for his continued support,
and Karla Fejfarova, Frantisek Malinka and Benoit
Piavaux for their expertise in bioinformatics.
REFERENCES
Baiker, M., Milles, J., Dijkstra, J., Henning, T.D., Weber,
A.W., Que, I., Kaijzel, E.L., Löwik, C.W., Reiber, J.H.,
Lelieveldt, B.P., 2010. Atlas-based whole-body
segmentation of mice from low-contrast Micro-CT
data. Med. Image Anal. 14(6). 723-37.
Bivand, R., Leisch, F., Maechler, M., 2015. Bitmap Images
(“Pixel Maps”). https://cran.r-
project.org/web/packages/pixmap.
Brown, J.M., Horner N.R., Lawson, T.N., Fiegel, T.,
Greenaway, S., Morgan, H., Ring, N., Santos, L.,
Sneddon, D., Teboul, L., Vibert, J., Yaikhom, G.,
Westerberg, H., Mallon, A.-M., 2018. A Bioimage
informatics platform for high-throughput embryo
phenotyping. Brief Bioinform. 19(1): 41-51
Bruker microCT, 2013a. Quantifying adipose tissue (fat) in
a mouse or rat by in-vivo microCT. 26 pp.
Bruker microCT, 2013b. Bone mineral density (BMD) and
tissue mineral density (TMD) calibration and
measurement by micro-CT using Bruker-MicroCT CT-
Analyser. 30 pp.
Ooms, J., 2018. Advanced Graphics and Image-Processing
in R. https://cran.r-project.org/web/packages/magick/
Paszke, A., Gross, S., Chintala, S., Chanan, G., Yang, E.,
DeVito, Z., Lin, Z., Desmaison, A., Antiga, L., Lerer,
A, 2017. Automatic differentiation in PyTorch. 31st
Conference on Neural Information Processing Systems
(NIPS 2017), Long Beach, CA, USA. 4 pp.
Pau, G., Fuchs, F., Sklyar, O., Boutros, M., Huber, W.,
2010. EBImage — an R package for image processing
with applications to cellular phenotypes.
Bioinformatics, 26(7), 979–981.
R Core Team, 2015. A language and environment for
statistical computing. R Foundation for Statistical
Computing, Vienna, Austria. http://www.R-
project.org/.
RStudio Team, 2016. RStudio: Integrated Development for
R. RStudio, Inc., Boston, MA URL
http://www.rstudio.com/.
Spoutil, F., Novosadova, V., Fejfarova, K., Malinka, F.,
Piavaux, B., Prochazka, J., 2018. MicroCT for high
throughput phenotyping: perspectives and obstacles.
Micro-CT User Meeting 2018. 187-190.
Wise, L.D., Winkelmann, C.T., Dogbas, B., Baqchi, A.,
2013. Micro-computed tomography imaging and
analysis in developmental biology and toxicology.
Birth Defects Res. C Embryo Today 99(2). 71-82.
microCT for Systematic Mouse Phenotyping
149
